AKDE provides a accurate, adaptive kernel density estimator based on the Gaussian Mixture Model for multidimensional data. This Python implementation includes automatic grid construction for arbitrary dimensions and provides a detailed explanation of the method.
pip install akde
from akde import akde
pdf, meshgrids, bandwidth = akde(X, ng=None, grid=None, gam=None)Performs adaptive kernel density estimation on dataset X.
X: Input data, shape(n, d), wherenis the number of samples anddis the number of dimensions.ng(optional): Number of grid points per dimension (default grid size based on dimensiond).grid(optional): Custom grid points for density estimation.gam(optional): Number of Gaussian mixture components. A cost-accuracy tradeoff parameter, wheregam < n. The default value isgam = min(gam or int(np.ceil(n ** 0.5)), n - 1). Larger values may improve accuracy but always decrease speed. To accelerate the computation, reduce the value ofgam.
pdf: Estimated density values on the structured coordinate grid of meshgrids (shape: (ng ** d,)). To reshape pdf back to match the original structured grid for visualization:pdf = pdf.reshape((ng,) * d)
meshgrids: Grid coordinates for pdf estimation (A list of d arrays, each of shape (ng,) * d)bandwidth: Estimated optimal kernel bandwidth (shape (d,))
import numpy as np
import matplotlib.pyplot as plt
from scipy.stats import norm
from akde import akde
# Set random seed for reproducibility
np.random.seed(42)
n_samples = 1000
# Define 5 Gaussian mixture components (means, standard deviations, and weights)
means = [-4, -2, 0, 2, 4] # Different means
stds = [0.5, 0.8, 0.3, 0.7, 1.0] # Different standard deviations
weights = [0.2, 0.15, 0.25, 0.2, 0.2] # Different weights (sum to 1)
# Generate samples from each Gaussian distribution
data = np.hstack([
np.random.normal(mean, std, int(n_samples * weight))
for mean, std, weight in zip(means, stds, weights)
]).reshape(-1, 1) # Reshape to (n,1) for AKDE
# Perform adaptive kernel density estimation
pdf, meshgrids, bandwidth = akde(data)
# Reshape the PDF to match the shape of the grid
pdf = pdf.reshape(meshgrids[0].shape)
# Compute the true density function by summing individual Gaussians
true_pdf = np.sum([
weight * norm.pdf(meshgrids[0], mean, std)
for mean, std, weight in zip(means, stds, weights)
], axis=0)
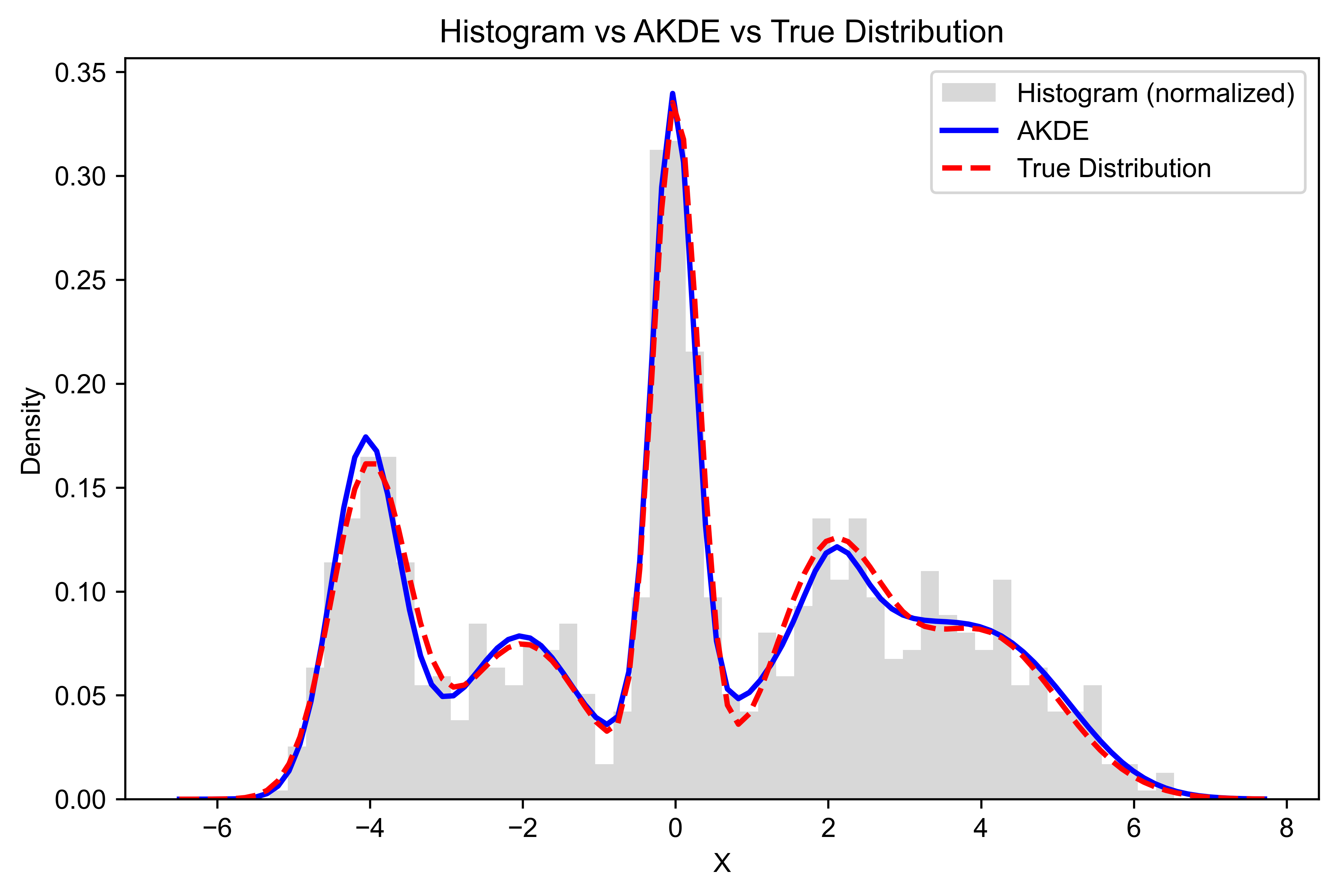
# Plot the density estimation, true distribution, and histogram
plt.figure(figsize=(8, 5))
plt.hist(data, bins=50, density=True, alpha=0.3, color='gray', label="Histogram (normalized)")
plt.plot(meshgrids[0], pdf, label="AKDE", color='blue', linewidth=2)
plt.plot(meshgrids[0], true_pdf, label="True Distribution", color='red', linestyle='dashed', linewidth=2)
plt.xlabel("X")
plt.ylabel("Density")
plt.title("Histogram vs AKDE vs True Distribution")
plt.legend()
plt.savefig("1D-AKDE-Density.png", transparent=False, dpi=600, bbox_inches="tight")
plt.show()import numpy as np
import matplotlib.pyplot as plt
from akde import akde
# Generate synthetic 2D mixture distribution data
np.random.seed(42)
n_samples = 1000
# Define mixture components
mean1, cov1 = [2, 2], [[0.5, 0.2], [0.2, 0.3]]
mean2, cov2 = [-2, -2], [[0.6, -0.2], [-0.2, 0.4]]
mean3, cov3 = [2, -2], [[0.4, 0], [0, 0.4]]
# Sample from Gaussian distributions
data1 = np.random.multivariate_normal(mean1, cov1, n_samples // 3)
data2 = np.random.multivariate_normal(mean2, cov2, n_samples // 3)
data3 = np.random.multivariate_normal(mean3, cov3, n_samples // 3)
# Combine data into a single dataset
X = np.vstack([data1, data2, data3])
# Perform adaptive kernel density estimation
pdf, meshgrids, bandwidth = akde(X)
# Reshape the PDF to match the shape of the grid
pdf = pdf.reshape(meshgrids[0].shape)
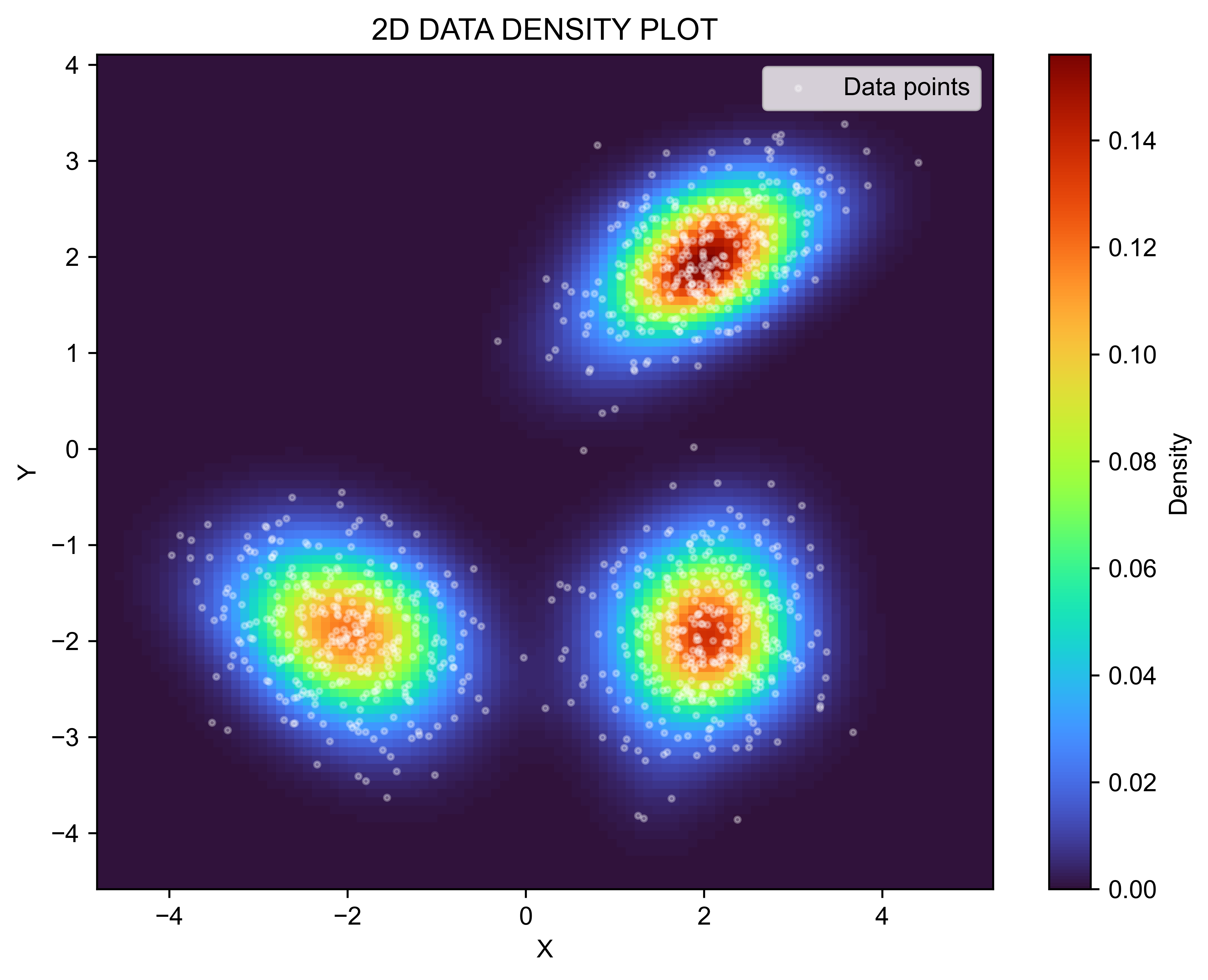
# Plot the density estimate
plt.figure(figsize=(8, 6))
plt.imshow(pdf, extent=[meshgrids[0].min(), meshgrids[0].max(),
meshgrids[1].min(), meshgrids[1].max()],
origin='lower', cmap='turbo', aspect='auto')
plt.colorbar(label="Density")
plt.scatter(X[:, 0], X[:, 1], s=5, color='white', alpha=0.3, label="Data points")
plt.xlabel("X")
plt.ylabel("Y")
plt.title("2D DATA DENSITY PLOT")
plt.legend()
plt.savefig("2D-AKDE-Density.png", transparent=False, dpi=600, bbox_inches="tight")
plt.show()import numpy as np
import plotly.graph_objects as go
from akde import akde
# Generate synthetic 2D mixture distribution data
np.random.seed(42)
n_samples = 10000
# Define mixture components
mean1, cov1 = [2, 2], [[0.5, 0.2], [0.2, 0.3]]
mean2, cov2 = [-2, -2], [[0.6, -0.2], [-0.2, 0.4]]
mean3, cov3 = [2, -2], [[0.4, 0], [0, 0.4]]
# Sample from Gaussian distributions
data1 = np.random.multivariate_normal(mean1, cov1, n_samples // 3)
data2 = np.random.multivariate_normal(mean2, cov2, n_samples // 3)
data3 = np.random.multivariate_normal(mean3, cov3, n_samples // 3)
# Combine data into a single dataset
X = np.vstack([data1, data2, data3])
# AKDE
pdf, meshgrids, bandwidth = akde(X)
Z = pdf.reshape(meshgrids[0].shape)
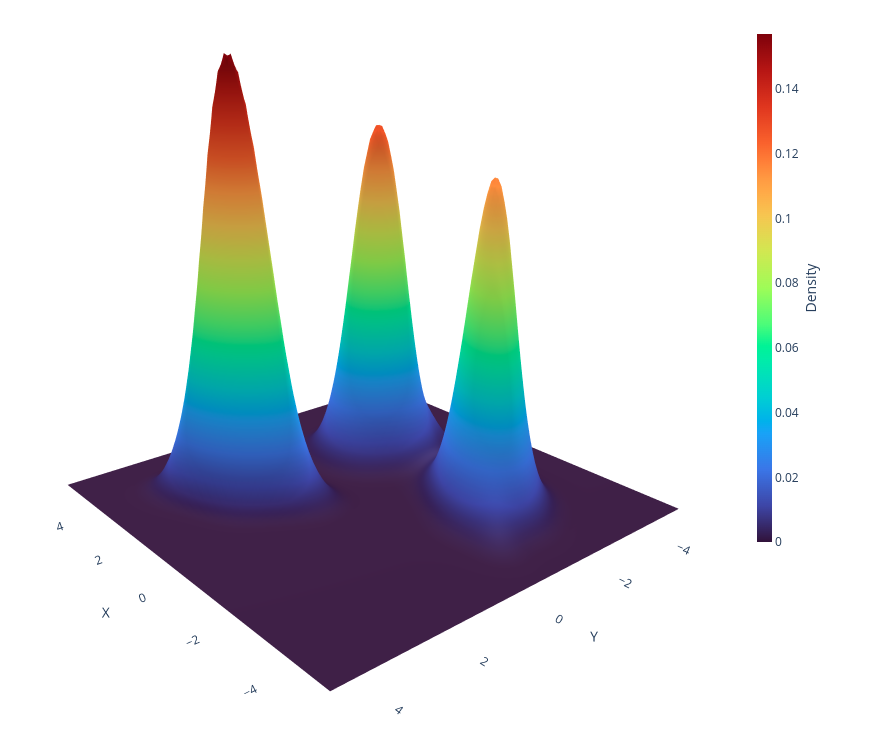
fig = go.Figure(data=[
go.Surface(
x=meshgrids[0], y=meshgrids[1], z=Z,
colorscale='Turbo',
opacity=1,
colorbar=dict(
title="Density",
titleside="right",
titlefont=dict(size=14),
thickness=15,
len=0.6,
x=0.7,
y=0.5
)
)
])
# Update layout settings
fig.update_layout(
title="",
scene=dict(
xaxis_title='X',
yaxis_title='Y',
zaxis_title="Density",
xaxis=dict(tickmode='array', tickvals=np.arange(-10, 11, 2.0), showgrid=False),
yaxis=dict(tickmode='array', tickvals=np.arange(-10, 11, 2.0), showgrid=False),
zaxis=dict(tickmode='array', tickvals=np.arange(0, 0.6, 0.1), showgrid=False),
),
margin=dict(l=0, r=0, t=0, b=0)
)
fig.update_layout(scene = dict(
xaxis = dict(
backgroundcolor="rgb(255, 255, 255)",
gridcolor="white",
showbackground=True,
zerolinecolor="white",),
yaxis = dict(
backgroundcolor="rgb(255, 255,255)",
gridcolor="white",
showbackground=True,
zerolinecolor="white"),
zaxis = dict(
backgroundcolor="rgb(255, 255,255)",
gridcolor="white",
showbackground=True,
zerolinecolor="white",),)
)
fig.update_scenes(xaxis_visible=True, yaxis_visible=True,zaxis_visible=False)
fig.update_layout(paper_bgcolor="rgba(0,0,0,0)", plot_bgcolor="rgba(0,0,0,0)", autosize=True)
fig.write_html("2D-AKDE-density-3D-projection.html")import numpy as np
import plotly.graph_objects as go
from akde import akde
# Set random seed for reproducibility
np.random.seed(12345)
# Number of samples
NUM = 10000
# Generate synthetic 3D data (mixture of 3 Gaussians)
means = [(2, 3, 1), (7, 7, 4), (3, 9, 8)]
std_devs = [(1.2, 0.8, 1.0), (1.5, 1.2, 1.3), (1.0, 1.5, 0.9)]
num_per_cluster = NUM // len(means)
# Create 2D numpy array data with shape: (NUM, 3)
data = np.vstack([
np.random.multivariate_normal(mean, np.diag(std), num_per_cluster)
for mean, std in zip(means, std_devs)
])
# AKDE
pdf, meshgrids, bandwidth = akde(data)
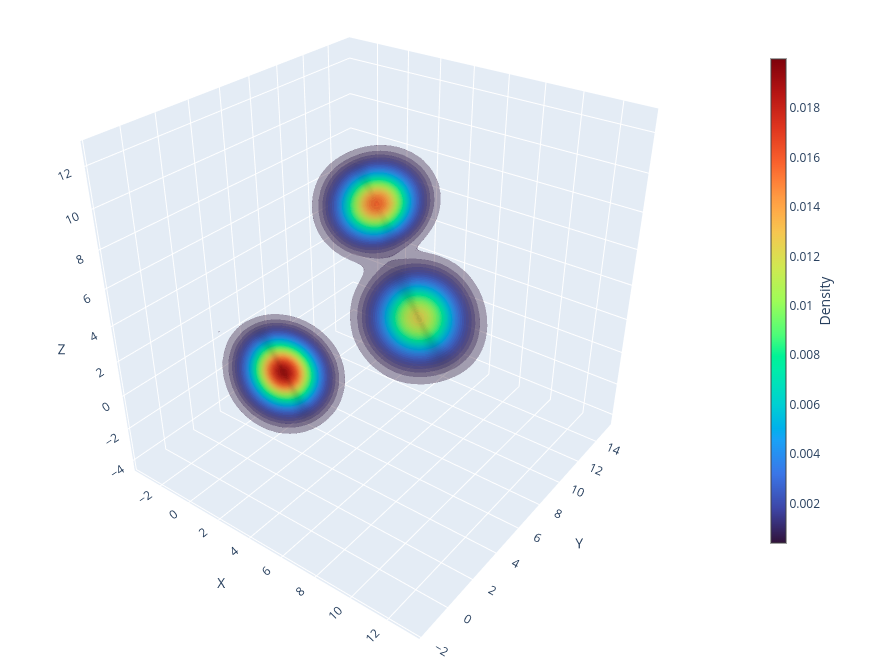
# ====== PLOTTING THE 3D DENSITY WITH PLOTLY VOLUME OR ISOSURFACE PLOT ======== #
fig = go.Figure()
fig.add_trace(
go.Volume(
x=meshgrids[0].ravel(),
y=meshgrids[1].ravel(),
z=meshgrids[2].ravel(),
value=pdf.ravel(),
isomin=pdf.max()/50,
opacity=0.2,
surface_count=50,
colorscale="Turbo",
colorbar=dict(
title="Density",
titleside="right",
titlefont=dict(size=14),
thickness=15,
len=0.6,
x=0.7,
y=0.5
)
)
)
# Layout customization
fig.update_layout(
scene=dict(
xaxis_title="X",
yaxis_title="Y",
zaxis_title="Z",
),
margin=dict(l=0, r=0, b=0, t=0),
autosize=True
)
# Export interactive plot in html, for export static figure install kaleido: pip install kaleido
fig.write_html("3d-density-akde-plotly.html")
# The 3D density plot from akde contains 128^3 points by default,
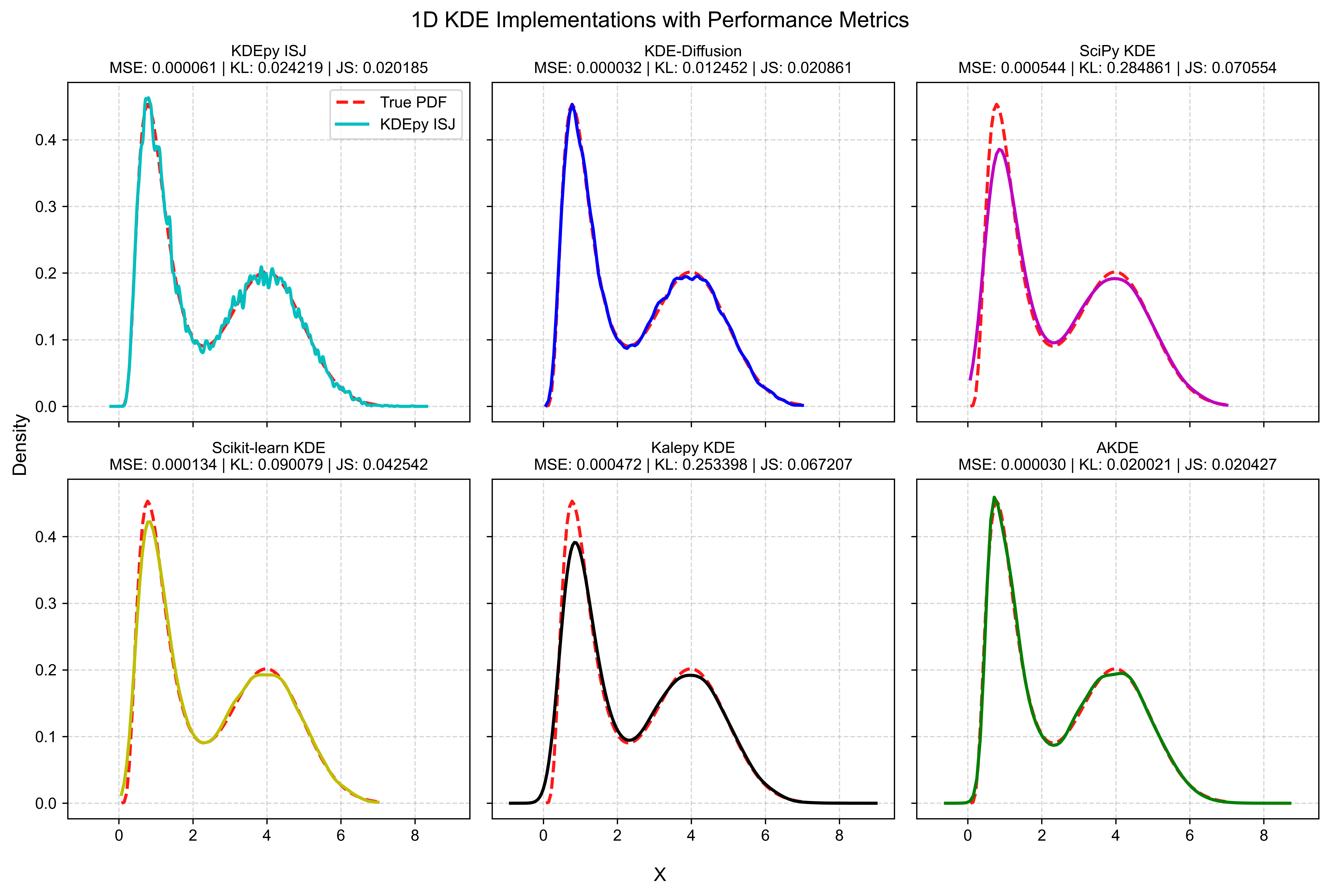
# it is not recommended to show Plotly interactive plot in ipynbBelow, we compare the performance of AKDE with various KDE implementations in Python by computing the Mean Squared Error (MSE), Kullback–Leibler (KL) divergence, and Jensen–Shannon (JS) divergence between the estimated density and the true distribution. Additionally, a goodness-of-fit test can be conducted using the SciPy one-sample or two-sample Kolmogorov-Smirnov test. For multidimensional cases, the fasano.franceschini.test package in R provides an implementation of the multidimensional Kolmogorov-Smirnov two-sample test.
- Probability density estimation (Galaxy Distribution, Stellar Population, distributions of temperature, wind speed, and precipitation, air pollutant concentration distribution...)
- Anomaly detection (Exoplanet Detection, characterization of rock mass discontinuities...)
- Machine learning preprocessing
This project is licensed under the MIT License - see the LICENSE file for details.
Contributions are welcome! Feel free to submit a pull request or open an issue.
Trung Nguyen
GitHub: @trungnth
Email: trungnth@dnri.vn
The original MATLAB implementation by Zdravko Botev does not seem to reference the algorithm described in the corresponding paper. Below, we present the mathematical foundation of the algorithm used in the AKDE method.
Kernel density estimation is a widely used statistical tool for estimating the probability density function of data from a finite sample. Traditional KDE techniques estimate the density using a weighted sum of kernel functions centered at the data points. The density is expressed as:
where
Despite its utility, traditional KDE suffers from a major challenge: the scaling efficiently to high-dimensional datasets aka The curse of dimensionality. The
Given a dataset
where
Here,
The parameters of the GMM—weights
where
The algorithm begins by normalizing the input data to fit within a unit hypercube
In the E-step, the algorithm computes the responsibilities
In the M-step, the model parameters are updated using the responsibilities:
These updates ensure that the log-likelihood of the data increases with each iteration. The EM process is implemented in the regEM function in AKDE, which terminates when the change in log-likelihood is below a predefined tolerance.
After optimizing the GMM parameters, the algorithm evaluates the estimated density function
where
These computations, implemented in the probfun function, are critical for the efficient and stable evaluation of the GMM.
A unique feature of AKDE is the dynamic adjustment of bandwidths for each Gaussian component based on the local curvature of the density. The curvature
where
The bandwidth
where
Entropy is used as a criterion to ensure the smoothness and generality of the density estimate. The entropy of the estimated density
The algorithm maximizes entropy iteratively, promoting a smooth and unbiased density estimate while avoiding overfitting. The AKDE implementation demonstrates the power of Gaussian Mixture Models in adaptive kernel density estimation. By combining the EM algorithm for parameter optimization, curvature-based bandwidth selection, and entropy maximization, the algorithm achieves flexible and accurate density estimation. The use of Cholesky decomposition ensures computational efficiency and stability, making the algorithm scalable for large datasets and high-dimensional problems. This seamless integration of mathematical rigor and computational efficiency highlights the versatility of GMMs in density estimation.